Cellpose
- Details
- deep-learning
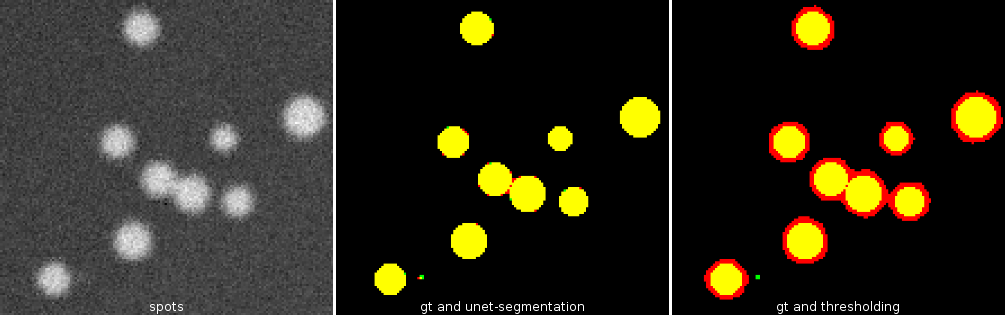
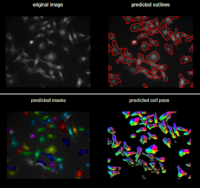
Cellpose allows to segment cells of different types from different imaging modalities. The deep-learning network is pre-trained with images of 70000 cells. Cellpose uses a vector flow representation of cells. The input is an image of the cells and optionally an image of stained nuclei.
MRI added a batch processing button, which allows to run the batch segmentation of cells from the graphical user interface instead of using the python from the command-line.
See also:
- Cellpose (Presentation for the MRI image analysis group, 05.02.2020)
- Cellpose reloaded (Presentation for the MRI image analysis group, 04.03.2021)